The research consortium contributes to the digital transformation of the drinking water sector by developing or further developing two software tools for continuous monitoring (I) and simulation (II) of organic and microbial drinking water quality parameters. The joint research project was funded within the German-Isreali Water technology Cooperation (Project-ID: 02WIL1553).
The three research groups from the Technical University of Hamburg, the Technion – Israel Institute of Technology, and the Technical University of Ilmenau, collaborated with industrial partners on their development. The software tools enable continuous assessment of current and future drinking water quality in the water distribution network, representing an innovation in terms of microbiological risk. Within the project, two tools were developed or enhanced regarding the monitoring and modeling of water quality parameters:
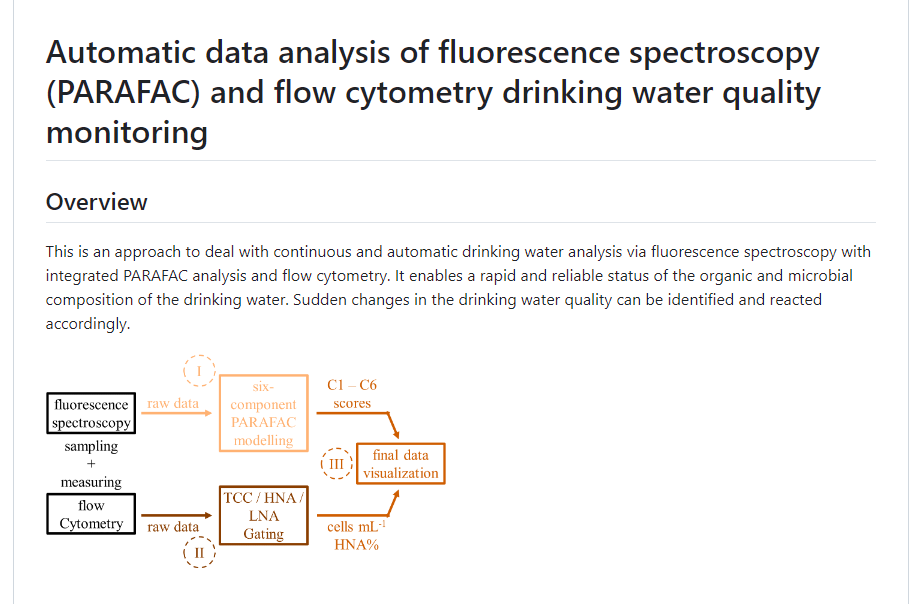
1. "Automized data analysis of fluorescence spectroscopy (PARAFAC) and flow cytometry drinking water quality monitoring"
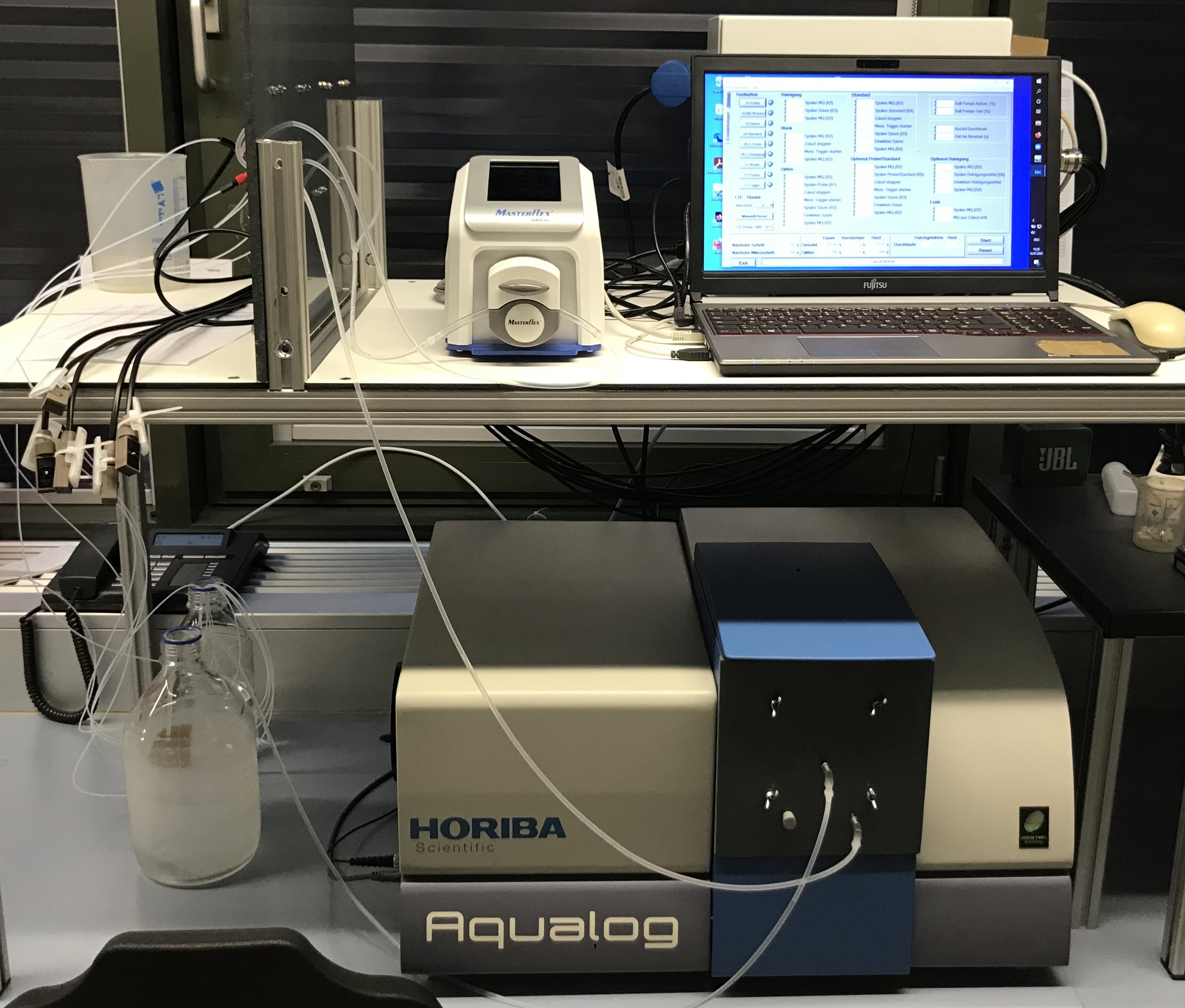
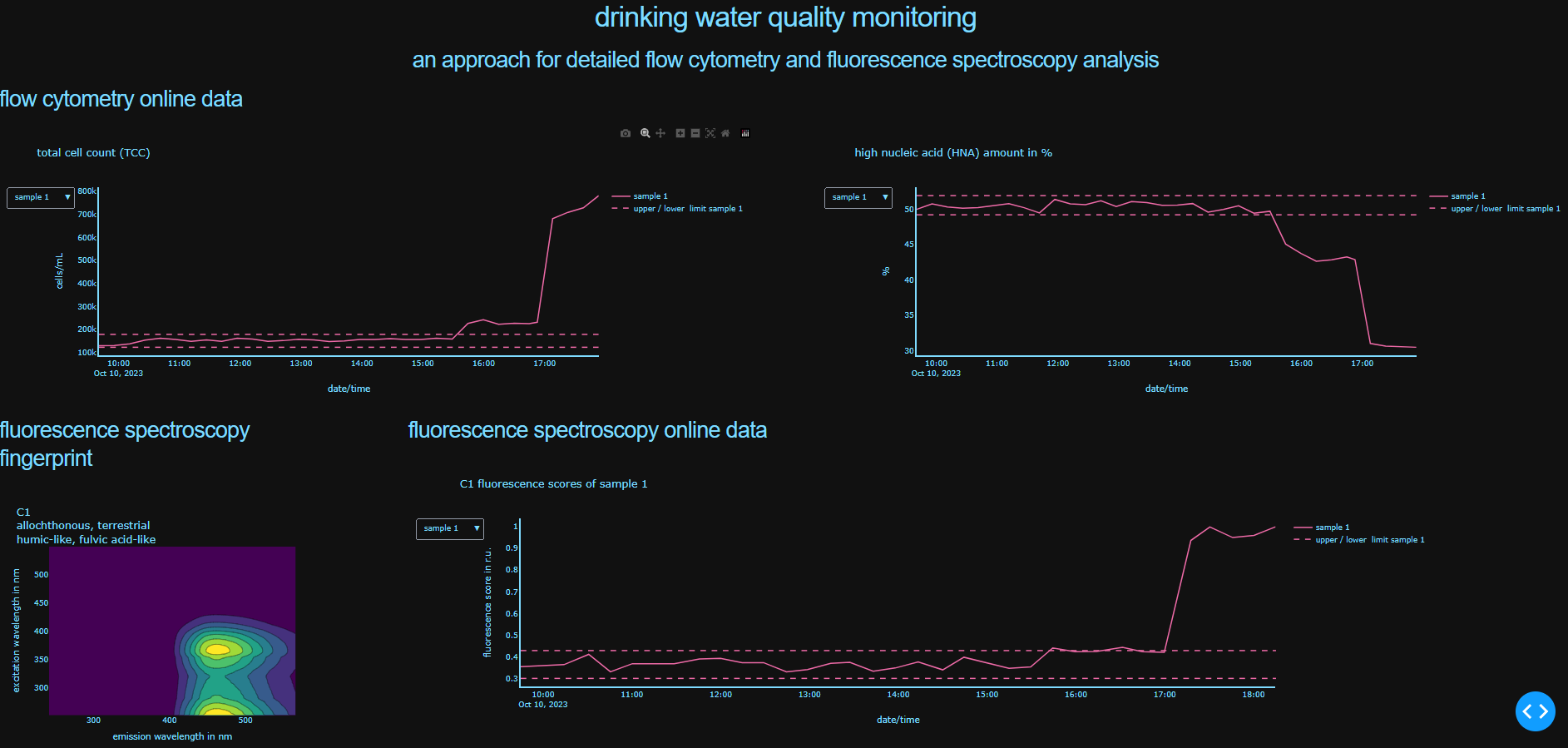
The developed approach allows fully automated parallel evaluation and visualization of continuously measured drinking water samples using fluorescence spectroscopy and flow cytometry. The established PARAFAC analysis is applied in an automated form to represent organic substances in the examined water through six organic fluorescence components. Additionally, water is characterized by cell the total cell count and the proportion of cells with relatively high nucleic acid content (HNA). Through visual representation in an interactive dashboard, changes in drinking water quality can be implemented in near real-time (15 min).
The software tool can be accessed in the media library under "Automated data analysis".
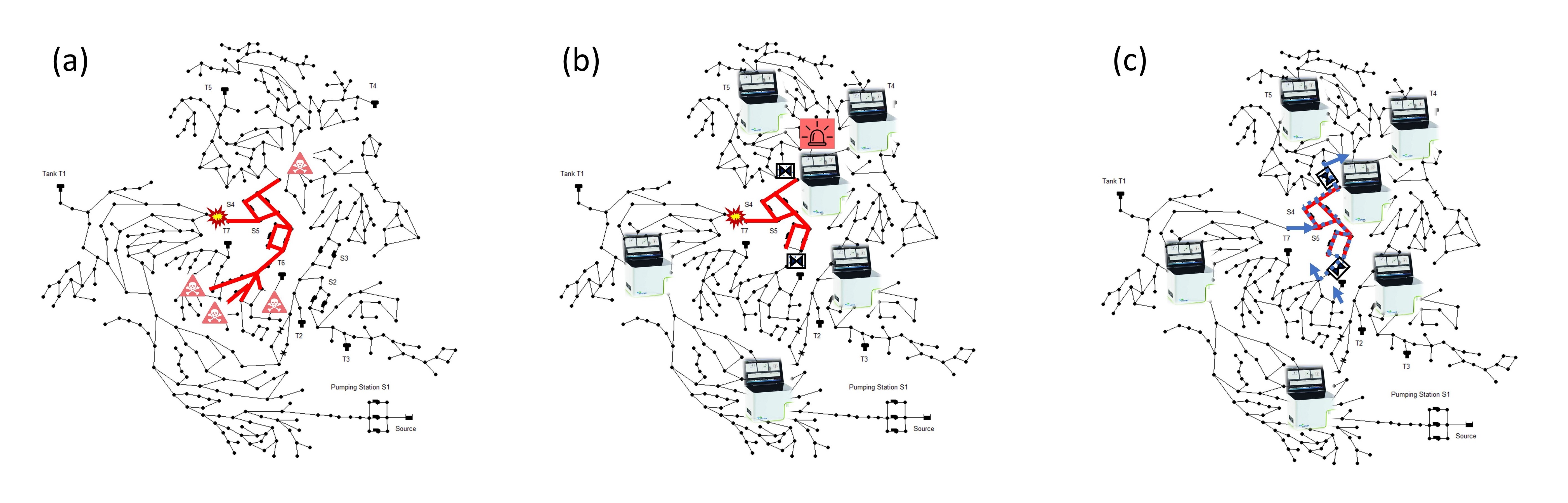
2. "EPyT-C – Fully independent multi-species reactive transport modeling extension for EPyT (EPANET-Python)"
The extension includes the ability to model the reactive transport of multiple substances (including pollutants), also known as multi-species reactive-transport (MSRT) modeling. This provides, through two developed modules, the ability to model free chlorine as well as the associated concentration of disinfection by-products such as trihalomethanes. The second module enables the modeling of recontamination in the drinking water based on the concentration of living bacteria.
The software tool can be accessed in the media library under "EPyT (EPANET-Python) - Independent multi-species reactive transport modelling extension".
Both software tools were successfully validated at the Technical University of Hamburg, and the Technion in laboratory and pilot scale and are freely available as open-access software via GitHub (see media library). Public availability allows:
· Direct implementation into existing drinking water networks with appropriate analytics.
· The possibility of individual customization or extension by specialized staff.